Structural Bioinformatics Group at Sapienza University of Rome
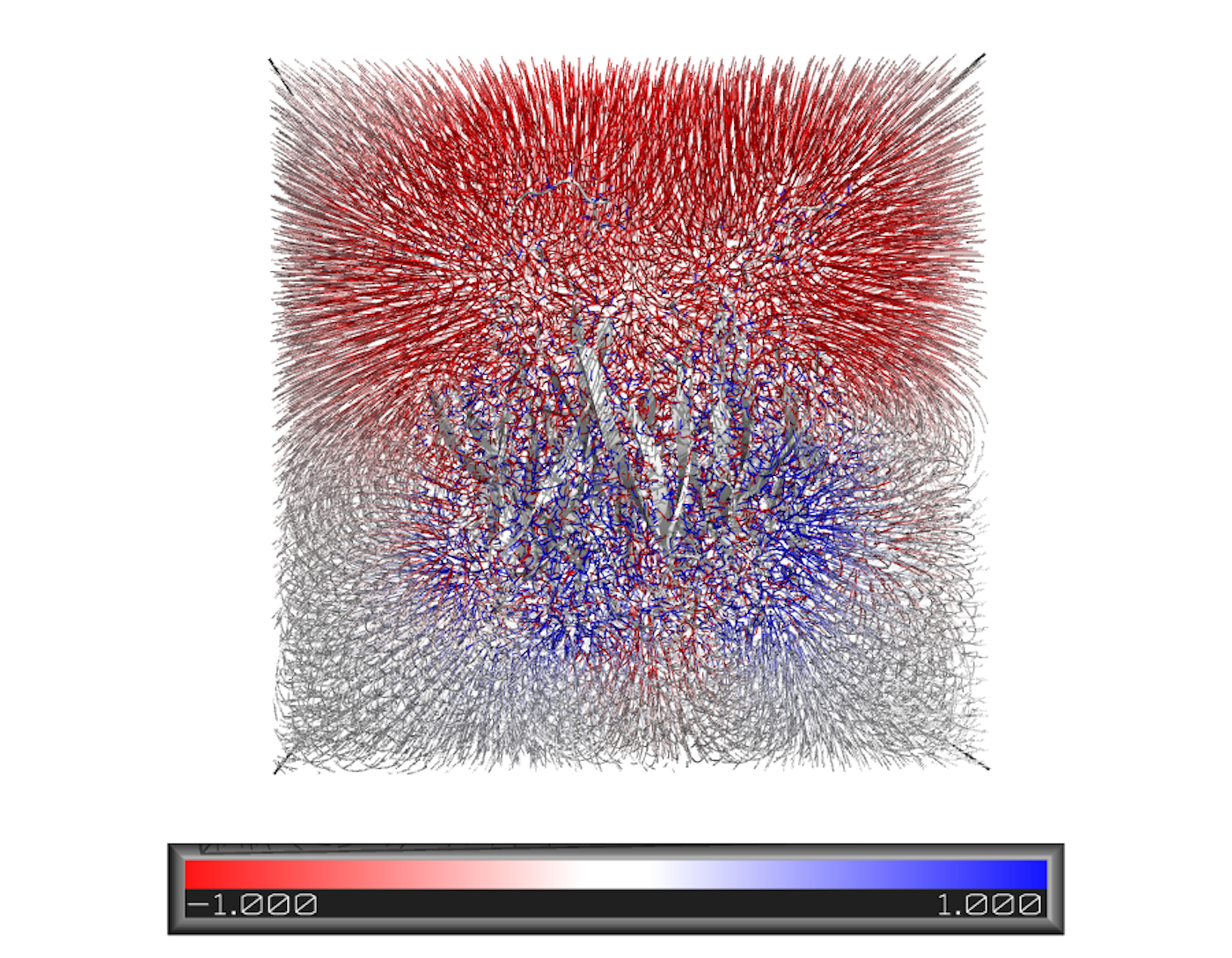
Structural Bioinformatics
We make use of computational methods to analyze and predict the three-dimensional structures of proteins and their interactions with potential drug molecules. This enables the design of more effective and targeted drugs.
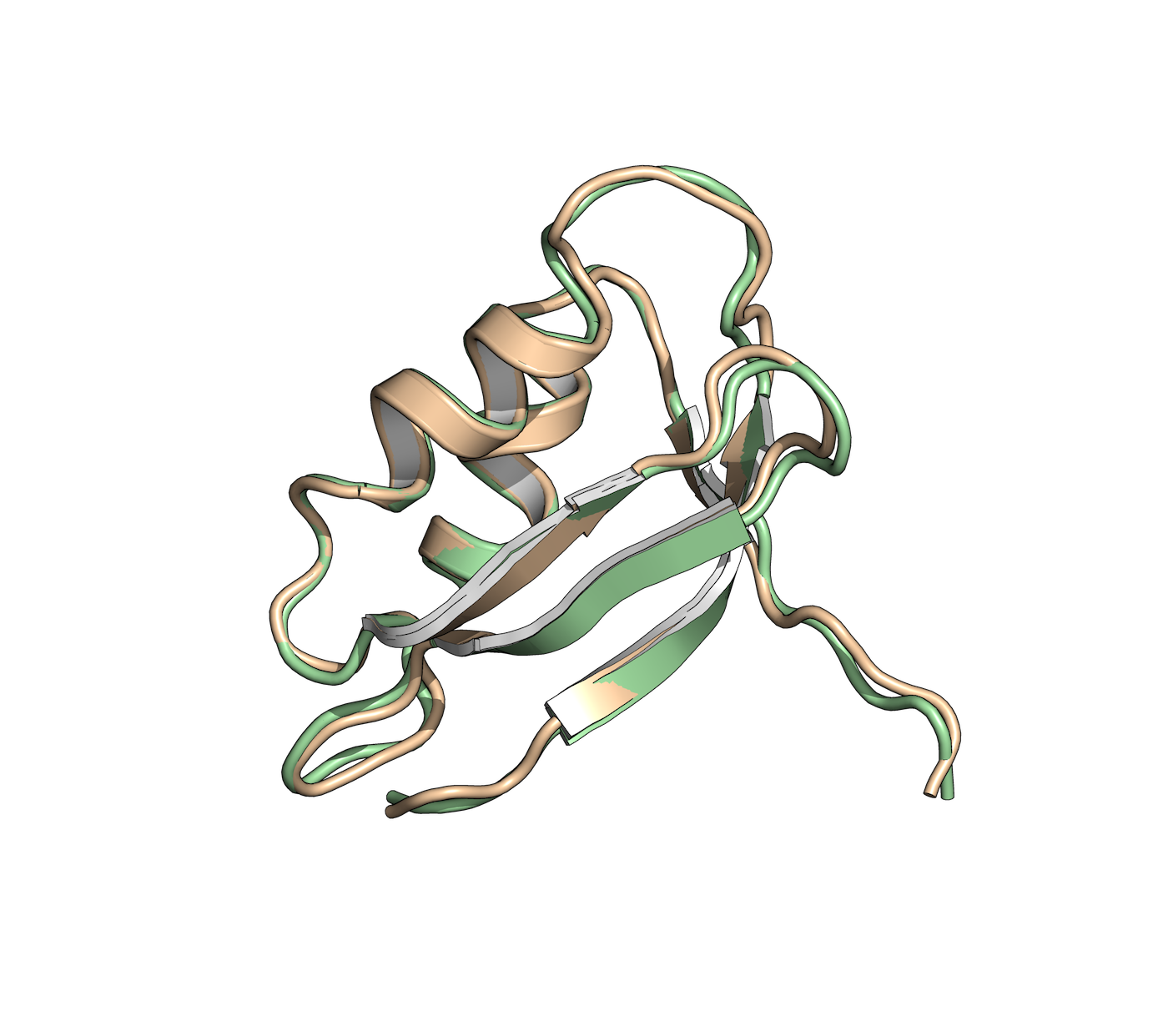
Protein Engineering
In the field of protein engineering, we help in the design and modification of proteins with improved properties, such as enhanced stability, activity, or specificity. This is achieved by studying the structure-function relationships of proteins and using computational tools to guide protein engineering strategies.
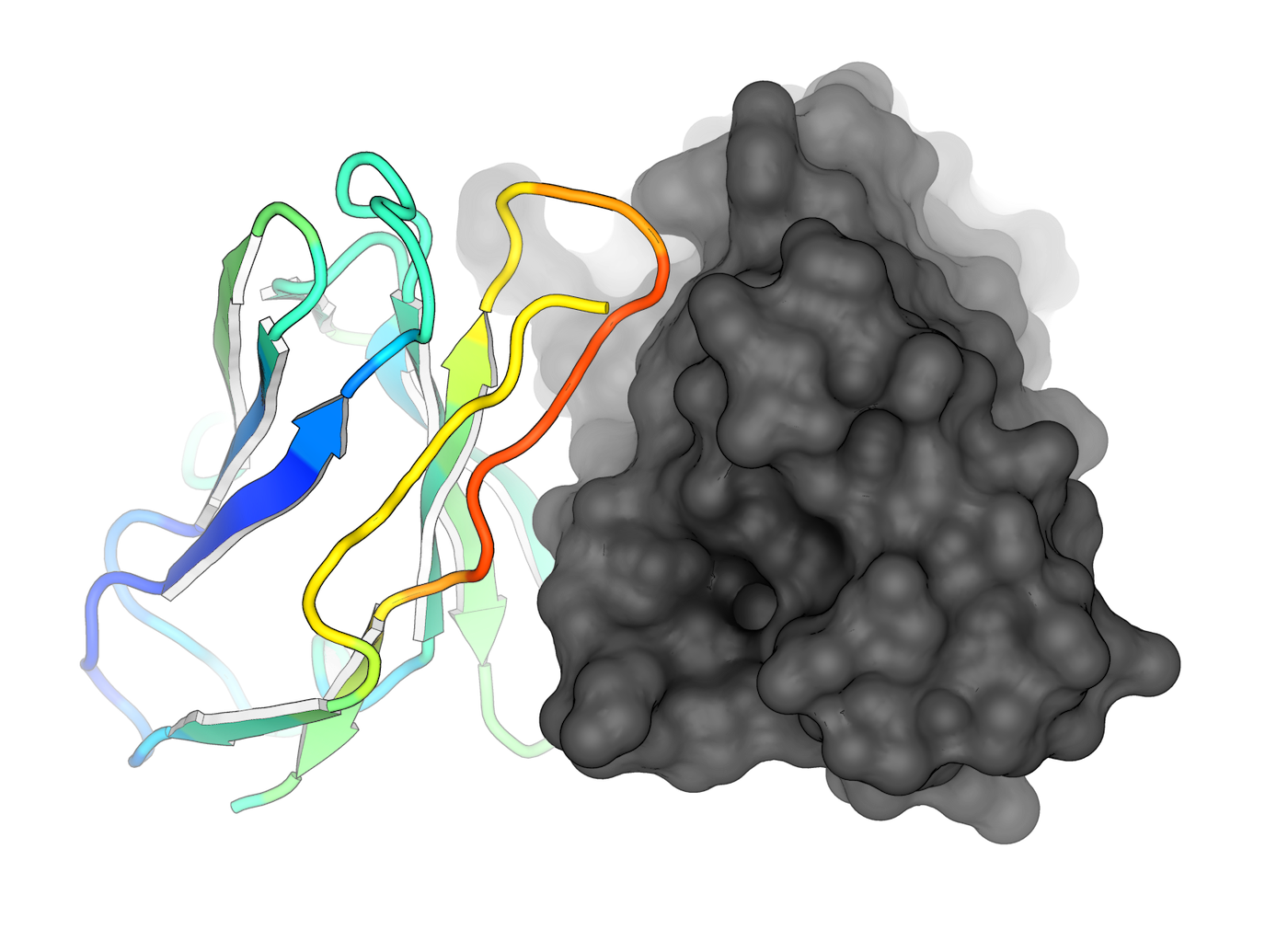
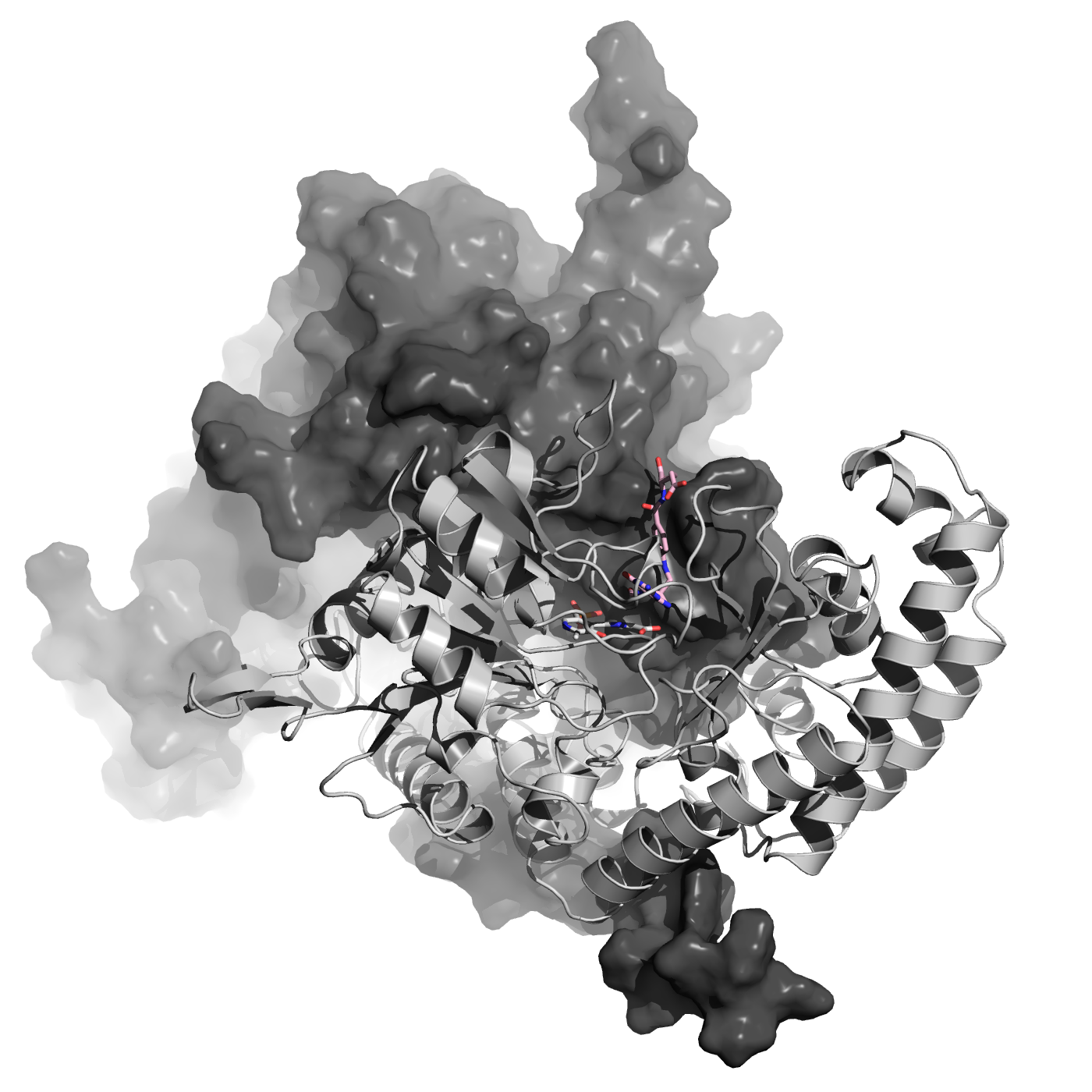
Protein Docking
We have expertise in protein docking, where we predict the binding modes and interactions between two or more proteins. This information is valuable for understanding protein-protein interactions, designing protein complexes, and studying biological pathways.
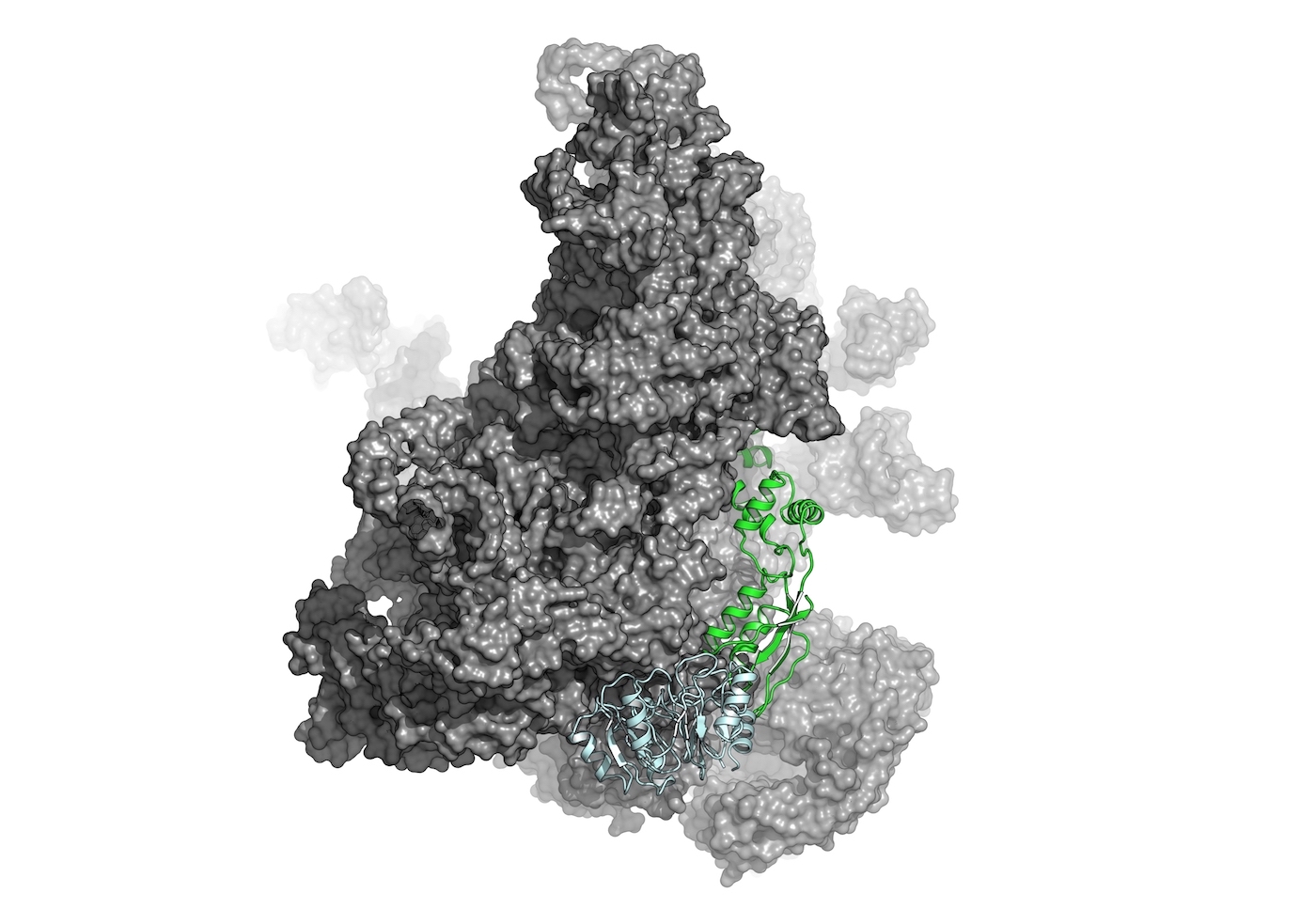
Functional Analysis
Structural bioinformatics plays a vital role in understanding the effects of genetic variations on protein structure and function. By analyzing the impact of mutations and genetic polymorphisms on protein structures, we can gain insights into the mechanisms underlying genetic diseases and develop personalized medicine approaches.
Algorithms and Software
Our Tools and algorithms in Python and R allow researchers to analyze and manipulate protein structures, enabling advanced analysis, modeling, and simulation of biomolecular systems.
Research
Our research is mainly focused on in silico Structural Bioinformatics and its in vitro and in vivo applications. Much of our research is disseminated in the form of articles, publicly available software, databases, and web-based servers.
PyMod
PyMod 3 is a PyMOL plugin, designed to act as a simple and intuitive interface between PyMOL and several bioinformatics tools (i.e., PSI-BLAST, Clustal Omega, MUSCLE, CAMPO, PSIPRED, and MODELLER). We are currently implementing also AlphaFold2 in PyMod3.
DockingPie
DockingPie is an open-source PyMOL plugin for assisting molecular and consensus docking analyses. DockingPie currently implements four docking engines, i.e., Smina, AutoDock Vina, ADFR, and RxDock, in an easy and highly intuitive way.
