Structural Bioinformatics Group at Sapienza University of Rome
Overview
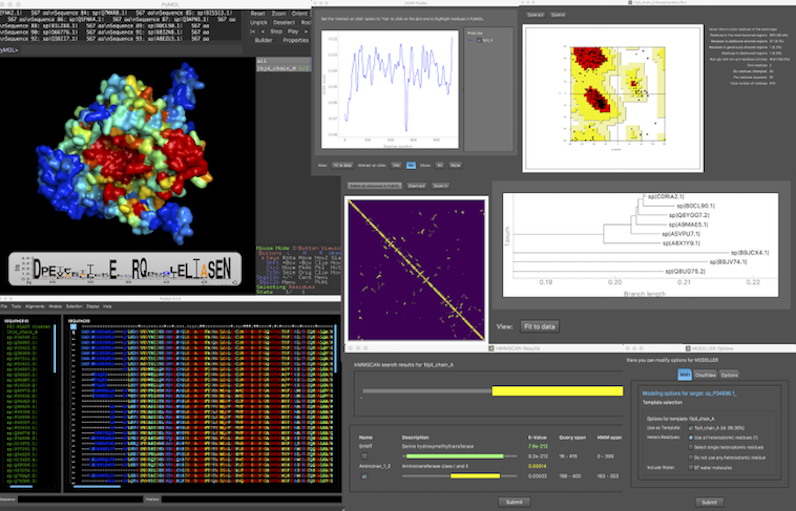
PyMod 3 is a PyMOL plugin, designed to act as simple and intuitive interface between PyMOL and several bioinformatics tools (i.e., PSI-BLAST, Clustal Omega, MUSCLE, CAMPO, PSIPRED, and MODELLER). The current PyMod release, PyMod 3, has been extended with a rich set of functionalities that substantially improve it over its predecessor (PyMod 2.0), particularly in its ability to build homology models through the popular MODELLER package.
Availability
PyMod 3 runs on Windows, macOS and Linux. It is compatible with PyMOL versions >= 2.3 and has been developed and tested on both incentive PyMOL builds (distributed by Schrodinger) and open source builds.
References
If you find PyMod useful for your research, please don't forget to cite it
PyMod Download
A series of introductory tutorials and accurate, in-depth information about all the PyMod 3 functionalities can be found in its User's Guide
Documentation is also available for PyMod3 at the GitHub site.
