Overview
Note
The SCR_FIND service is not available anymore on this server. We are sorry for the inconvenience. However we implemented it in PyMod 3, so you may still use it in within PyMod.
Description
The Structurally Conserved Regions (SCRs) are protein segments that conserve the same main-chain conformation in all the three-dimensional structures analysed, excluding the intervening regions whose structure differs markedly amongst different proteins. The SCRs were presumably subjected to similar constraints during the divergent evolution of a family or superfamily of proteins from a common ancestor; therefore they possibly contain most of the determinants necessary to maintain the fold. SCR_FIND is a method for the automatic identification of the SCRs, which assigns a score for each position of a multiple structure alignment based on the mean RMSD computed for the correspondent coordinates of the structurally equivalent alpha carbons.
Reference
Paiardini, A., Bossa, F., and Pascarella, S. (2005). CAMPO, SCR_FIND and CHC_FIND: a suite of web tools for computational structural biology. Nucleic Acids Res. 33, W50-55. PMID: 15980521
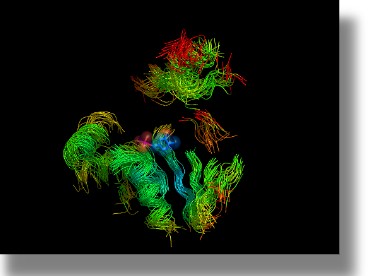
Superimposition of the SCRs found in PLP, fold type I enzymes. The backbones of 23 superposed structures is shown as solid oval ribbon. Seventeen regions with a mean positional RMSD = 3.0 Å, lacking insertions and deletions were detected and the corresponding coordinates coloured according to the RMSD value. PLP is displayed as slate CPKs. Oxygen atoms are coloured red, nitrogen atoms blue, and phosphorus purple.
