Note
The CAMPO web service is not available anymore on this server. However, we recently integrated the CAMPO algorithm in PyMod 3. If you want to use CAMPO, you may use the version implemented in PyMod.
Overview
Description
The importance of a residue for maintaining the structure and function of a protein can usually be inferred from how conserved it appears in a multiple sequence alignment of that protein and its homologues. CAMPO is a tool useful for the identification of conserved amino-acid sites among sequence homologues. Conserved amino-acid sites were presumably subjected to similar constraints during the divergent evolution of a family or superfamily of proteins from a common ancestor; therefore they possibly contain most of the determinants necessary to maintain the fold and function of a protein. CAMPO is a method for the automatic identification of these sites, which assigns a score for each position of a multiple sequence alignment based on the scores of a mutational matrix, the sequence identity between sequences being compared and, optionally, the distance between clustered conserved residues.
Reference
Paiardini, A., Bossa, F., and Pascarella, S. (2005). CAMPO, SCR_FIND and CHC_FIND: a suite of web tools for computational structural biology. Nucleic Acids Res. 33, W50-55. PMID: 15980521
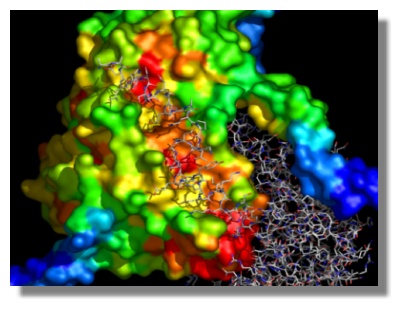
Mapping of evolutionary conservation on the molecular surface of serine hydroxymethyltransferase from H. sapiens. The results obtained are expressed in units of Standard Deviations from the mean conservation value. Dark blue corresponds to maximal variability, red to maximal conservation. The monomer-monomer interface involving the interaction between a surface cleft and the N-terminal arm of the other monomer is shown to highlight the evolutionary conservation-structural function relationship.
